Traditional fragment-based drug discovery approaches have revolutionized drug design by focusing on small, low-affinity molecules that can efficiently explore chemical space. In recent years, the development of 3D-fragment libraries has emerged as a powerful method to enhance the success rate of hit identification and lead optimization. This article delves into the design principles and applications of 3D-fragment libraries, exploring their potential in identifying novel drug candidates and accelerating the drug discovery process. By leveraging structural information and advanced computational tools, these libraries offer valuable insights into target-binding preferences and aid in the rational design of more potent and selective therapeutics.
Introduction:
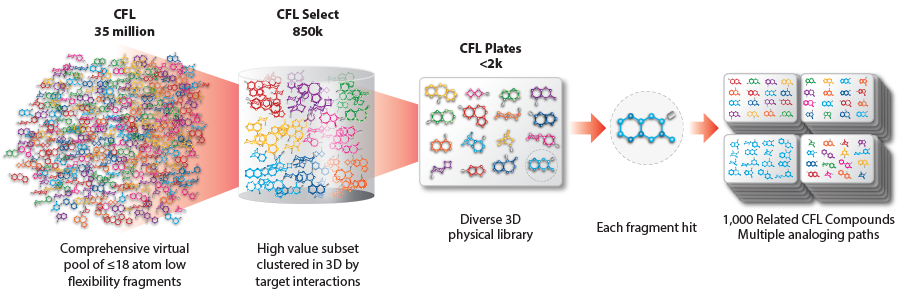
Fragment-based drug discovery has gained prominence as an efficient approach to find lead compounds, focusing on small fragments that interact with a target protein or receptor. Traditional fragment libraries have primarily relied on 2D chemical space exploration, often lacking the specificity required for successful lead identification and optimization. In contrast, 3D-fragment libraries harness the power of structural information to create a diverse set of fragments that can efficiently sample the spatial and chemical features of target binding sites.
Design Principles:
The design of a 3D-fragment library involves careful selection of fragment compounds that possess both chemical and spatial diversity. Chemoinformatics tools and computational methods enable the exploration of chemical space, ensuring a broad coverage of physicochemical properties and structural motifs. Additionally, considerations such as fragment size, flexibility, and scaffold diversity guide the selection process. Chemical synthesis and fragment linking strategies are employed to generate larger and more potent compounds from initial hits and identify lead candidates for further optimization.
Structural Information and Computational Tools:
Leveraging structural information is vital in designing a 3D-fragment library. X-ray crystallography, NMR spectroscopy, and computational modeling techniques provide valuable insights into target binding sites, allowing for rational fragment selection and library generation. Molecular docking, fragment fingerprinting, and virtual screening techniques can also be employed to predict fragment binding modes and prioritize compounds for experimental testing. These computational tools complement experimental efforts, enhancing the efficiency of hit identification and lead optimization.
Applications:
3D-fragment libraries have shown promising applications in various target classes and therapeutic areas. They enable the exploration of protein-protein interactions, allosteric sites, and complex target binding pockets that are challenging for traditional small molecule libraries. By screening against diverse protein targets, these libraries aid in the discovery of new chemical starting points and improve hit rates and chemical space coverage. The insights gained from 3D-fragment libraries can accelerate lead optimization and improve the quality of drug candidates, increasing the chances of success in clinical trials.
Advantages and Future Perspectives:
The design of 3D-fragment libraries offers several advantages over traditional fragment-based approaches. By considering 3D spatial information, these libraries have the potential to identify fragments with higher affinity, selectivity, and binding modes. Moreover, the systematic exploration of chemical and structural diversity expands the range of viable drug targets. The continuous evolution of structural biology and computational techniques presents exciting opportunities for further improving the design and utility of 3D-fragment libraries. Additionally, collaborations and knowledge-sharing among researchers can enhance the accessibility and applicability of these libraries in drug discovery efforts.
Conclusion:
The design of 3D-fragment libraries marks a significant advancement in fragment-based drug discovery, offering new avenues for identifying potent and selective drug candidates. By leveraging structural insights and computational tools, these libraries provide a comprehensive exploration of chemical space, facilitating hit identification and lead optimization. As the field continues to evolve, 3D-fragment libraries hold immense potential in accelerating the drug discovery process and expanding the repertoire of therapeutic options for complex diseases.